问题描述
我正在尝试做一个以前看起来很简单的操作,但是我没有在网络上找到明确的解决方案。
我有这种桌子:
tibble(
block = c(1,1,2,2),tag = letters[1:6],start = c(15,54,78,27,45,80),end = c(50,80,90,40,76,100),direction = c(-1,-1,1),anchor = c(FALSE,TRUE,FALSE,FALSE)
) -> df1
# A tibble: 6 x 6
block tag start end direction anchor
<dbl> <chr> <dbl> <dbl> <dbl> <lgl>
1 1 a 15 50 -1 FALSE
2 1 b 54 80 -1 TRUE
3 1 c 78 90 1 FALSE
4 2 d 27 40 1 FALSE
5 2 e 45 76 1 TRUE
6 2 f 80 100 1 FALSE
我在block列中有组,每个组只有1个anchor。
给定anchor == TRUE,如果锚点方向为direction * -1(-1 ,则需要反转(direction[anchor] == -1))块内的坐标,也需要保留锚点坐标(start和end ,并调整anchor == FALSE的另一个坐标和坐标,以使其保持新月形,但比例不变(长度和距离和下游标签)。
为简化起见,如果组的锚点为-1,则需要重新调整坐标。
这意味着,如果anchor == -1则:
-
ancho * -1 - 标记订单必须还原
- 将更改坐标,并保持标签的长度以及它们之间的距离相同
然后,输出只需要像这样:
tibble(
block = c(1,tag = c("c","b","a","d","e","f"),start = c(44,84,end = c(56,119,FALSE)
) -> df2
# A tibble: 6 x 6
block tag start end direction anchor
<dbl> <chr> <dbl> <dbl> <dbl> <lgl>
1 1 c 44 56 -1 FALSE
2 1 b 54 80 1 TRUE
3 1 a 84 119 1 FALSE
4 2 d 27 40 1 FALSE
5 2 e 45 76 1 TRUE
6 2 f 80 100 1 FALSE
如下所示,长度和配对距离保持不变:
df1 %>%
group_by(block) %>%
mutate(
TagDistance = lead(start) - end,len = end - start
)
# A tibble: 6 x 8
# Groups: block [2]
block tag start end direction anchor TagDistance len
<dbl> <chr> <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
1 1 a 15 50 -1 FALSE 4 35
2 1 b 54 80 -1 TRUE -2 26
3 1 c 78 90 1 FALSE NA 12
4 2 d 27 40 1 FALSE 5 13
5 2 e 45 76 1 TRUE 4 31
6 2 f 80 100 1 FALSE NA 20
df2 %>%
group_by(block) %>%
mutate(
TagDistance = lead(start) - end,len = end - start
)
# A tibble: 6 x 8
# Groups: block [2]
block tag start end direction anchor TagDistance len
<dbl> <chr> <dbl> <dbl> <dbl> <lgl> <dbl> <dbl>
1 1 c 44 56 -1 FALSE -2 12
2 1 b 54 80 1 TRUE 4 26
3 1 a 84 119 1 FALSE NA 35
4 2 d 27 40 1 FALSE 5 13
5 2 e 45 76 1 TRUE 4 31
6 2 f 80 100 1 FALSE NA 20
图形表示是这样:
library(ggplot2)
library(gggenes)
df1 %>%
ggplot(aes(xmin = start,xmax = end,y = as.factor(block),forward = direction,fill = anchor)) +
geom_gene_arrow() +
geom_gene_label(aes(label = tag)) +
theme_genes()
#
df2 %>%
ggplot(aes(xmin = start,fill = anchor)) +
geom_gene_arrow() +
geom_gene_label(aes(label = tag)) +
theme_genes()
预先感谢
解决方法
我解决了,很愚蠢,也许有更好的解决方法?
tibble(
block = c(1,1,2,2),tag = letters[1:6],start = c(15,54,78,27,45,80),end = c(50,80,90,40,76,100),direction = c(-1,-1,1),anchor = c(FALSE,TRUE,FALSE,FALSE)
) -> a
a
# A tibble: 6 x 6
block tag start end direction anchor
<dbl> <chr> <dbl> <dbl> <dbl> <lgl>
1 1 a 15 50 -1 FALSE
2 1 b 54 80 -1 TRUE
3 1 c 78 90 1 FALSE
4 2 d 27 40 1 FALSE
5 2 e 45 76 1 TRUE
6 2 f 80 100 1 FALSE
然后,我按block分组,并进行了许多启发式的算术运算,例如:
a %>%
group_by(block) %>%
mutate(
anchor_direction = direction[anchor],position_relative_to_anchor = case_when(
anchor ~ NA_character_,(start < start[anchor]) | (start == start[anchor] && end < end[anchor]) ~ "upstream",start > start[anchor] ~ "downstream"
),TagDistance = if_else(
position_relative_to_anchor == "upstream",start[anchor] - end,start - end[anchor]
),length = end - start,newstart = case_when(
anchor ~ start,anchor_direction == 1 ~ start,position_relative_to_anchor == "upstream" ~ end[anchor] + TagDistance,position_relative_to_anchor == "downstream" ~ start[anchor] - TagDistance
),newend = case_when(
anchor ~ end,anchor_direction == 1 ~ end,position_relative_to_anchor == "upstream" ~ newstart + length,position_relative_to_anchor == "downstream" ~ newstart - length
),start = case_when(
anchor ~ start,position_relative_to_anchor == "upstream" ~ newstart,position_relative_to_anchor == "downstream" ~ newend
),end = case_when(
anchor ~ end,position_relative_to_anchor == "upstream" ~ newend,position_relative_to_anchor == "downstream" ~ newstart
)
) %>%
arrange(block,start,end) %>%
mutate(
direction = direction * anchor_direction
) %>%
select(
-c(
anchor_direction,position_relative_to_anchor,TagDistance,length,newstart,newend
)
) -> a
a
# A tibble: 6 x 6
# Groups: block [2]
block tag start end direction anchor
<dbl> <chr> <dbl> <dbl> <dbl> <lgl>
1 1 c 44 56 -1 FALSE
2 1 b 54 80 1 TRUE
3 1 a 84 119 1 FALSE
4 2 d 27 40 1 FALSE
5 2 e 45 76 1 TRUE
6 2 f 80 100 1 FALSE
最后,我将其与预期结果进行了比较:
tibble(
block = c(1,tag = c("c","b","a","d","e","f"),start = c(44,84,end = c(56,119,FALSE)
) -> b
setdiff(a,b)
# A tibble: 0 x 6
# Groups: block [0]
# … with 6 variables: block <dbl>,tag <chr>,start <dbl>,end <dbl>,direction <dbl>,anchor <lgl>
欢迎任何更好的解决方案。



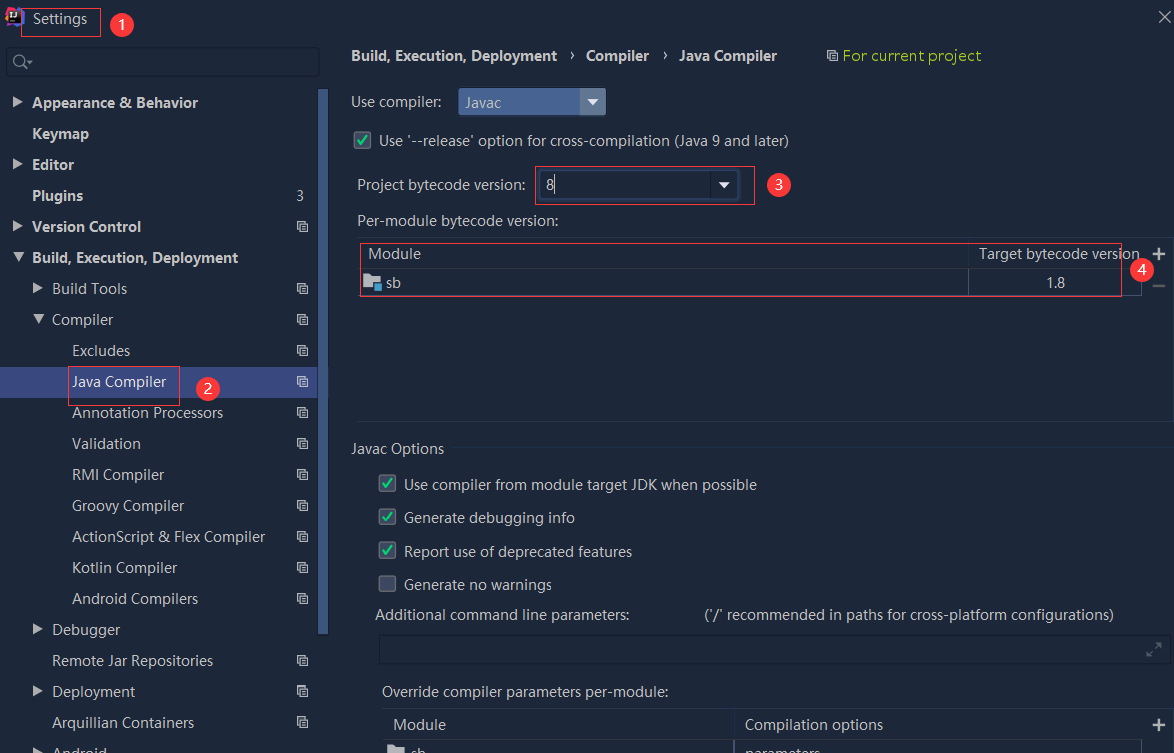 依赖报错 idea导入项目后依赖报错,解决方案:https://blog....
依赖报错 idea导入项目后依赖报错,解决方案:https://blog....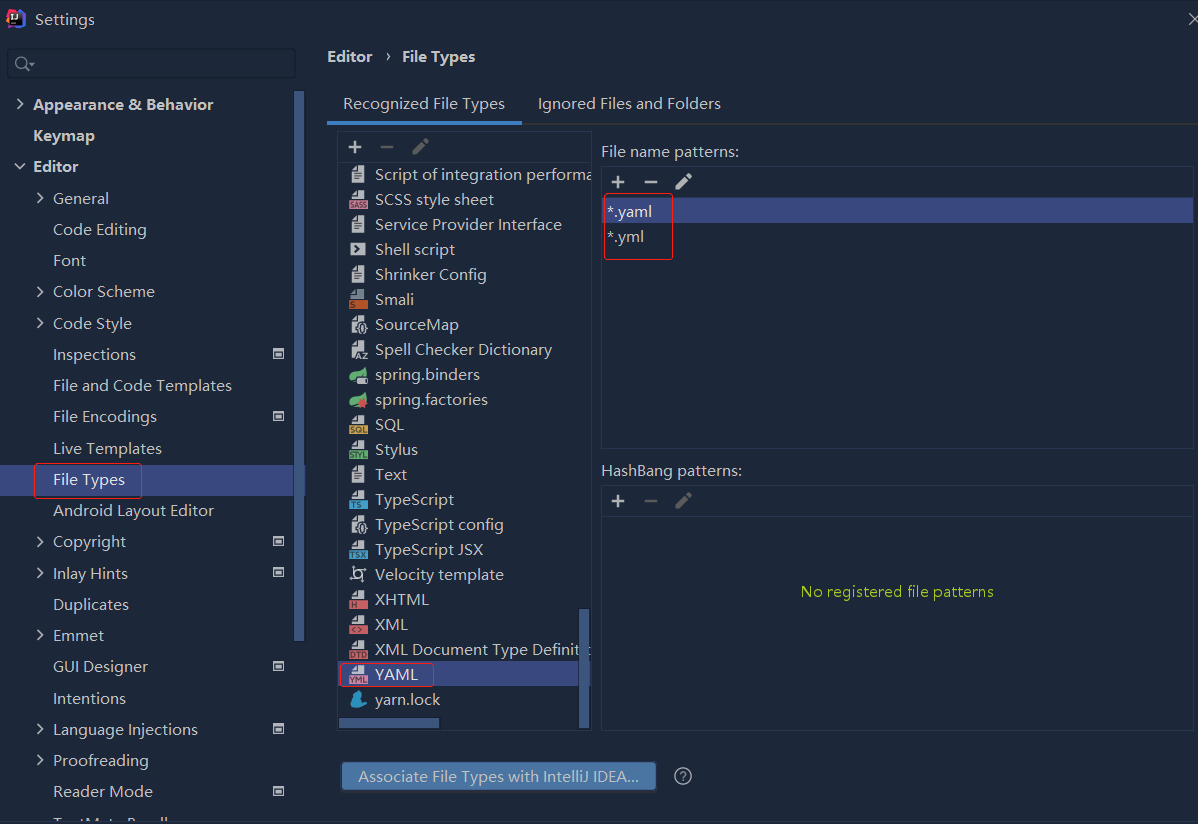
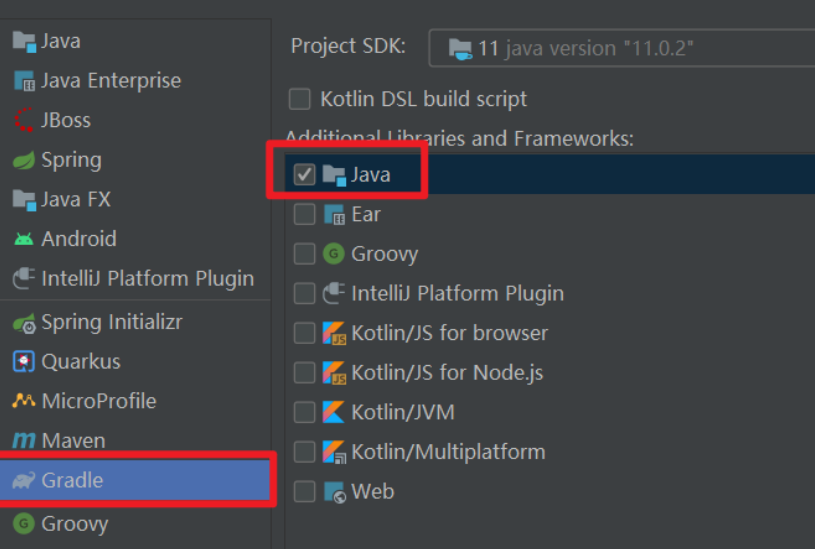 错误1:gradle项目控制台输出为乱码 # 解决方案:https://bl...
错误1:gradle项目控制台输出为乱码 # 解决方案:https://bl...